Denmark’s Minkstermination
The decision announced by the Danish prime minister, Mette Frederiksen, on November 4th to destroy all minks in the northern country is projected to result in the destruction of more than 15 million minks. The Danish government claimed their decision was supported by evidence that a mutated variant of SARS-CoV-2 spread among minks had infected humans and that this variant could interfere with future vaccine effectiveness. However, the severity of the mink viral strain is still under debate. Slaughtering all minks nationwide, with its far reaching implications for the whole fur industry, has been described as both aggressive and overly cautious.
While causing surprise all over the world and leading to concerns of the severity of the astonishing move, the spread of SARS-CoV-2 among farmed mink did not happen overnight. The first reported cases of COVID among farmed minks in Denmark occurred on June 17th and led to more than 5,000 minks being euthanized. On September 18th, Denmark’s infectious disease research institute, the Statens Serum Instituton, alerted that the mutated virus from mink had formed trans-infection chains in humans and warned that these virus mutations could attenuate community immunity. Urged to take immediate action to minimize the risk of wider spreading of the new virus mutant, on November 4th the Danish prime minister announced the order to kill all minks nation-wide.
Strain tracing and variant calling
One natural question to ask is how virus strains are being traced, especially for mutations that might have unwanted potential to boost transmission and/or lethality. Examples from an earlier study1 focusing on SARS Coronavirus-2 variant tracing during the early outbreak of COVID-19 patients in northern Germany illustrate bioinformatics’ role in virus strain tracing during this pandemic. Specifically, the techniques applied include variant calling and comparative analysis of single nucleotide polymorphisms (SNPs). Genomic data was collected via metagenomic RNA-sequencing and amplicon-sequencing before subsequent variant calling identified frequently observed clusters of SNPs. The mutations that enhanced the replication and transmission of the virus are more likely to take precedence and therefore serve as signatures for identifying popularized strains (Figure 1). Strain clusters are critical for evaluating the dynamics of virus mutation, for diagnostic and therapeutic purposes as well as to inform policies aiming to contain the spread of the virus.
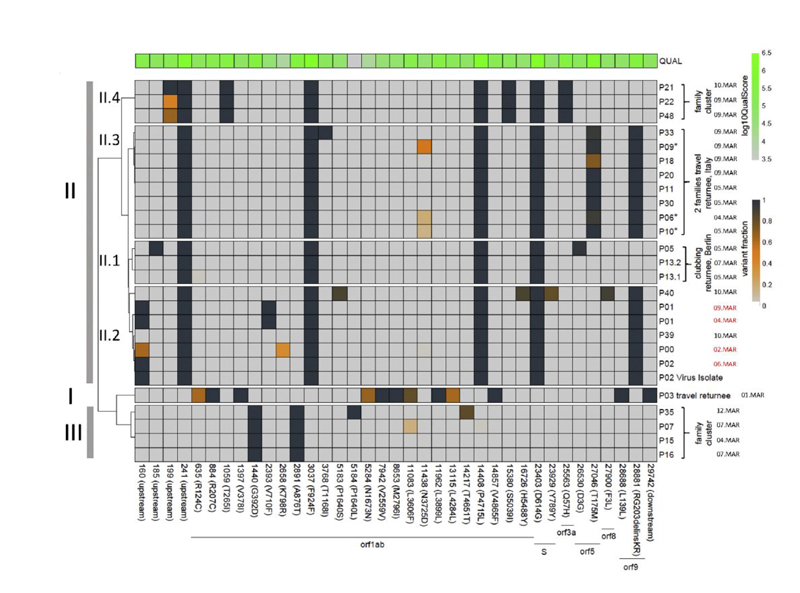
Figure 1.
Clustering of viral variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequences recovered from the index patient, patient 1, patient 2 and 19 SARS-CoV-2 sequences from respiratory swabs collected at the same time period in comparison to the reference sequence, NC_045,521. Nucleotide positions are indicated at the bottom. Only variants with sufficient coverage (>10) and single nucleotide polymorphism (SNP) present in more than 33% of all reads and in at least one sample are included. I-III summarizes sequence patterns as defined by SNPs. The frequency of variants is indicated by the heat map ranging from grey (reference), yellow to dark blue (variant). The quality score per individual site is indicated at the top. * indicates members within one family. Sampling dates are indicated on the right with the sampling date of the cases in the index cluster labelled in red.
Challenges
1. Potential Overinterpretation
Comparative genomics has been a common practice in detecting similarities among and differences between samples. However, one bigger challenge following variant discovery is evaluating the significance or potential impact of SARS-Cov-2 mutations on pandemic containment efforts. Scientists express skepticism upon the claim that mutations in one of the clusters identified from Denmark’s farm minks could attenuate the efficacy of vaccines under active development. More scrutinized trials are needed to evaluate the risks of the newly popularized strain interfering with vaccination efforts.
2. Lack of universal guidelines
Risky incidents of intra- and interspecies transmissions of SARS-Cov-2 have been captured and spotlighted. However, universal guidelines are still needed to prevent interspecies transmission between humans and other natural hosts. In similar emergencies, detailed criteria will be immeasurably valuable for supporting or halting such extreme public health measures as euthanizing large herds of animals. Despite enormous costs, overcaution can still be greatly beneficial, when well guided.
Reference
1. Pfefferle, S., Günther, T., Kobbe, R., Czech-Sioli, M., Nörz, D., Santer, R., Oh, J., Kluge, S., Oestereich, L., Peldschus, K. and Indenbirken, D., 2020. SARS Coronavirus-2 variant tracing within the first Coronavirus Disease 19 clusters in northern Germany. Clinical Microbiology and Infection.
Published: November 18, 2020

