The ANCHOR pipeline
ANCHOR is a high-resolution metagenomics pipeline, the result of a multi-disciplinary collaboration between C3G and researchers at Institut de recherche en biologie végétale (IRBV) at Université de Montreal. Published in 2019 in the journal Environmental Microbiology, the pipeline demonstrated unprecedented accuracy in its characterization of microbiome samples [1]. Dr. Emmanuel Gonzalez, co-creator of the pipeline and the metagenomics specialist at C3G was motivated by his observation that existing tools were not performing well on real-life datasets. “C3G’s strategy towards microbiome analysis had been to run commonly used pipelines, but the resolution would sometimes deliver foggy results that made the interpretation challenging. When I joined C3G, another pipeline was buzzing, using a machine learning algorithm to remove sequence noise – a characteristic of metagenomics samples. That was quite the thing back in 2015. It was clever, light and incredibly fast, except… I happened to realize that its accuracy was dropping significantly with real-world experiments involving multiple samples.
In designing ANCHOR, Dr Gonzalez and co-creator Dr. Nicholas Brereton had resolved that the tool would be written with a strong focus on understanding the biology of metagenomics experiments. Dr Gonzalez recalls that “before I even began writing down any code, we sat down together and set up some ground rules for this new pipeline, based on the respect towards the inherent complexity of any biological system and a reassessment of the capacity and ability of technical advances to handle biological complexity. I think this starting point is what made ANCHOR stand out amidst similar metagenomic pipelines. For example, our first rule went absolutely against the trend: let’s not modify sequences!
Confined habitats
In the publication introducing ANCHOR, Drs Gonzalez and Brereton reanalysed metagenomic data from surface swabs within the International Space Station (ISS). The ISS experiment had originally used the Qiime pipeline and the conclusions of the analysis were that no differences were present when comparing the surface microbial ecosystems of the Destiny (US laboratory) and Harmony (crew sleeping quarters) modules. The reanalysis using ANCHOR substantially improved the scale of data capture as well as the accuracy and resolution of the findings, providing microbial classifications at the level of individual species. The reanalysis not only led to exciting novel discoveries regarding the ISS environment but also fundamentally changed the major conclusion of the experiment, with significant differences clearly identified between the modules (Figure 1). These significant differences detected by ANCHOR included increases in microbiome bacteria associated with the laboratory animals within the Destiny module, such as Helicobacter typhlonius, a species endemic to rodent research laboratories on earth [1].
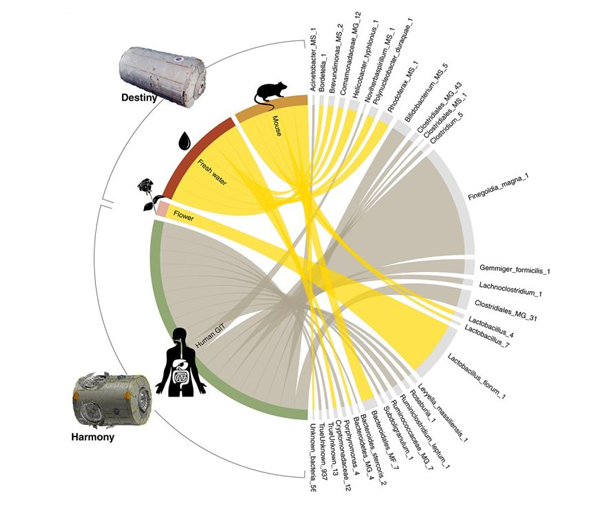
Figure 1. ISS Destiny and Harmony Module differential abundance. Significantly differentially abundant taxa, their ISS location and illustrations indicating their known associations.
Presented as part of the scientific article, ANCHOR was used to reanalyse metagenomic data from surface swabs within the International Space Station (ISS). The experiment originally used the Qiime pipeline and the conclusions of the analysis were that no differences were present when comparing the surface microbial ecosystems of the Destiny (US laboratory) and Harmony (crew sleeping quarters) modules. The reanalysis substantially improved the scale of data capture as well as the accuracy and resolution of the findings, now at microbial species-level. “The researchers from the first analysis did a great job, but the pipeline they used limited their ability to analyse what was inside the with precision” adds Emmanuel. The reanalysis not only led to exciting novel discoveries regarding the ISS environment but also fundamentally changed the major conclusion of the experiment, with significant differences clearly identified between the modules (Figure 1). These significant differences included increases in microbiome bacteria associated with the laboratory animals within the Destiny module, such as Helicobacter typhlonius, a species endemic to rodent research laboratories on earth(1).
Human microbiome health
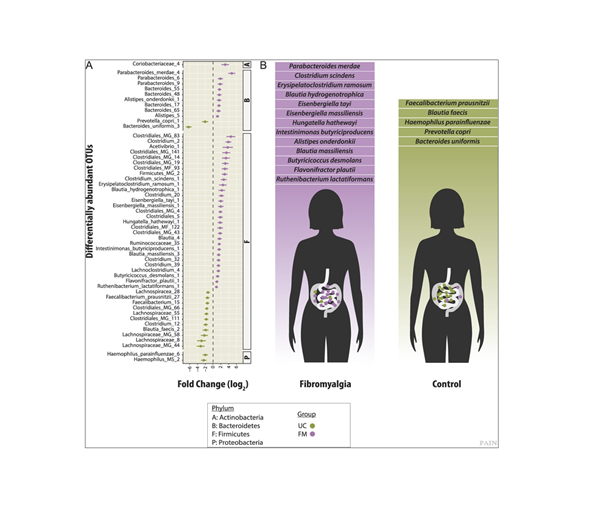
The ANCHOR pipeline was designed to improve data accuracy in high complexity real-world systems, and its applicability quickly moved from the ISS to terrestrial health concerns. “We transitioned this approach directly to the field of human health later in 2019 after Dr. Amir Minerbi, a medical doctor from the Alan Edwards Pain Management Unit, showed great interest to use our pipeline to characterise the bacterial microbiota of women suffering of fibromyalgia”, says Emmanuel. That collaboration led to the discovery of the first microbiome association to chronic pain by comparative analysis of the gut microbiomes of healthy women with fibromyalgia patients [2]. In patients with fibromyalgia, significant increases of species such as Clostridium scindens and Butyricicoccus desmolans, which are bottlenecks in secondary bile production (via 7α-dehydrogenase activity) as well as bacterial derived androgens (20α- and 20β-hydroxysteroid dehydrogenase activity) which may interact with the endocrine system (Figure 2). These are important discoveries in the field which has sparked a new research direction in the pain field and the research has already been recognised around the world.
Long-term Space Travel
At the end of 2019, the team behind ANCHOR responded to a grant call made by the Canadian Space Agency to promote projects on Health & Life Science data and sample mining. They proposed to analyse astronaut microbiomes using improved metagenomics to study the impact of long-term space travel upon astronaut health. “We’re grateful to the Canadian Space Agency for giving us the chance to apply high resolution microbiome analysis within the space sciences again and particularly excited to see whether there is an observable impact of long term confinement on the microbiome health of astronauts.
Les microbes de la Station spatiale internationale
1. Gonzalez E, Pitre F, Brereton N. ANCHOR: A 16S rRNA gene amplicon pipeline for microbial analysis of multiple environmental samples. Environmental Microbiology. 2019.
2. Minerbi A, Gonzalez E, Brereton NJ, Anjarkouchian A, Dewar K, Fitzcharles M-A, et al. Altered microbiome composition in individuals with fibromyalgia. Pain. 2019.
ANCHOR: a 16S rRNA gene amplicon pipeline for microbial analysis of multiple environmental samples
Read more
Learn more aboutANCHOR
Author: Emmanuel Gonzalez
Science Quebec article:Read now





